Volume 11 - Year 2024 - Pages 51-61
DOI: 10.11159/jbeb.2024.007
Investigating Sex-Dependent Differences in Tensional Homeostasis at the Single-Cell and Tissue Levels
Nourhan El Sherif1, Diya Desai1, Josephine Dunphy1, Marin Perez1, Sabrina Wilderotter1, Michael Smith1, Brianne Connizzo1
1Department of Biomedical Engineering, Boston University
44 Cummington Mall, Boston, MA, USA
nourhane@bu.edu; ddesai08@bu.edu; josied@bu.edu; maripere@bu.edu; swild@bu.edu; msmith@bu.edu; connizzo@bu.edu
Abstract - Sex-dependent variances in cellular and tissue mechanics are poorly understood despite evidence showing females' increased susceptibility to soft tissue injuries and degenerative diseases compared to that of males. This project outlines a collaborative effort between the Smith and Connizzo Labs to explore sex-specific differences in cellular mechanosensing, focusing on how cells respond to external stimuli to maintain a resting level of mechanical stress (tensional homeostasis) and extracellular matrix (ECM) remodeling. Using traction force microscopy (TFM) and a novel tissue bioreactor system, this study aims to quantify differences in tensional homeostasis between male and female tendon cells (tenocytes) and investigate strain transfer through the ECM in mouse flexor digitorum longus (FDL) tendons. The process involves both cellular and tissue components. At the cellular level, male and female mouse tenocytes are derived through digestion, seeded onto polyacrylamide gels, and then imaged to determine the coefficient of variation in traction forces. On the tissue level, tenocyte nuclei and cellular membranes are stained, loaded into a confocal-mounted bioreactor, stretched, and imaged. Results reveal efficient traction force experimentation setup with challenges in cell culture viability, but successful staining protocols for live tissue imaging have been obtained with images from male and female tendons subjected to strains of 0%, 3%, 6%, and 9%. Using these images, subsequent MATLAB image analysis calculated nuclear strain transfer, nuclear strain, and cellular strain for both sexes. The study provides quantitative measures of sex-dependent differences in tissue mechanosensing mechanisms, carrying potential implications for gender-specific therapeutics.
Keywords: Sex-dependent differences, Tensional homeostasis, Biomechanics, Soft tissue injuries.
© Copyright 2024 Authors This is an Open Access article published under the Creative Commons Attribution License terms. Unrestricted use, distribution, and reproduction in any medium are permitted, provided the original work is properly cited.
Date Received: 2024-07-21
Date Revised: 2024-09-26
Date Accepted: 2024-10-15
Date Published: 2024-12-06
1. Introduction
Tensional homeostasis is the ability of cells and tissues to respond to external stimuli. Homeostasis of the ECM is an essential part of organ development and wound healing, as well as a necessary part of maintaining a balance of production and breakdown of the ECM [1]. Disruption of this homeostasis can lead to soft tissue damage and degenerative diseases such as osteoarthritis, osteoporosis, sarcopenia, and carpal tunnel syndrome [2-5]. Additionally, current research posits that biological sex differences in tissue repair and regeneration correlate to higher risk of these diseases in females [6-7]. While the ECM structure and content regarding sex-based differences have been identified, the effects of cell signaling have not [8-10]. The lack of understanding how cell signaling affects homeostasis of the ECM creates a barrier of progress for modulation of ECM remodeling, which restricts enhancement for tissue healing. Therefore, the significance of this study is its novelty in mechanobiology– it is a new and unexplored area for exploring sex differences. This is important because identifying sex-based differences in regulatory pathways leads to better understanding of ECM remodeling modulation, a necessary step for improving representative research in therapeutic treatments for degenerative diseases. Current research on remodeling-related structural and organization components has uncovered the importance of collagen regulation; however, many scientists believe further research is necessary to fully understand ECM remodeling within sex-based differences [11-12]. Our project addresses these questions by quantifying a cell’s ability to regulate tensional homeostasis and understanding sex-based regulatory pathways with a bioreactor capable of exploring cellular responses to mechanical stimuli, allowing for in-vitro experiments. Therefore, the research provides necessary scientific knowledge to develop more advanced treatments for degenerative diseases that utilize ECM remodeling.
While studies have shown sex discrepancy in the incidence of human tendon injuries [13-14], there is a lack of studies that have investigated the fundamental variations between male and female cells in their ability to regulate tensional homeostasis. This study uses tenocytes, specialized tendon cells responsible for ECM secretion and accumulation [15]. Notably, there is a scarcity of comprehensive traction data concerning tenocytes, with only two known studies quantifying cytoskeletal and nuclear strains in this specific cell type [16-17]. By conducting this research study, we aimed to bridge a gap in knowledge, providing an understanding of the quantitative difference between male and female mechanical regulation, contractility, and homeostasis.
The Connizzo Lab has already found that age has a significant influence on mechanical stress response in tendon cells, with aged explants exhibiting a “less robust response” characterized by properties like decreased DNA content, cell density, and cell activity. It was also observed that some age-related differences were sex-dependent, implying that both age and sex have a factor in maintaining tensional homeostasis. The male tenocytes reportedly favored matrix degeneration, apoptosis, and remodeling while females aimed for tissue preservation distinguished by decreased inflammation and decreased activity [18]. This information, coupled with the fact that female mice had superior Achilles tendon healing compared to male mice and ovariectomized females, clarifies that male and female tenocytes are inherently distinct in responding to mechanical stress [19].
In this project, we first used TFM to measure cell prestress and tensional homeostasis. This process, developed in the Smith Lab, involves creating a micropatterned polyacrylamide (PAA) gel, seeding tenocytes on that gel, and imaging the resulting pattern after the cell applies contractile force to displace the patterned dots [20]. Once this new, displaced pattern is run through a MATLAB code, the traction force vectors and coefficients of variation (CVs) are calculated. The difference in male and female CVs can then be tested for significance with a two-sample t-test.
From the data acquired in TFM experiments, we aimed to determine a significant difference in how male and female tenocytes maintain tensional homeostasis. This difference is quantified by calculating the CV of the traction field for male and female tenocytes, where a lower CV characterizes a cell closer to homeostasis. In knowing the CV of both sexes, therapeutic developers and researchers can set to analyze the biomechanical cause of the difference, leading to further research in patient-specific therapies to account for differences in homeostasis maintenance and keeping more patients out of risk of disease progression and even cancer [21]. This experimentation can then be conducted using a bioreactor to assess how tendons maintain tensional homeostasis when stretched.
At the tissue level, this study used a novel bioreactor designed to apply uniaxial strain to tissues. This allowed for the direct application of mechanical load onto tissue, specifically the FDL tendon, enabling the study of mechanically driven cell behavior by placing uniaxial strain on a tendon for a designated period of time. Notably, the bioreactor simultaneously allows for real-time imaging along with mechanical loading, thus enabling the quantification of deformation and understanding strain transfer through the ECM.
To place mechanical stresses on the tenocytes, a custom-made bioreactor was used to assess the strain transfer with MATLAB computational analysis. The 2020-2021 Boston University Senior Design Team developed the hardware of the bioreactor, which is compatible with the Olympus FV3000 scanning confocal microscope and contains a 250g load cell (Honeywell AL311AR). It allows for visualization of cells within a tendon explant.
Finally, the collaboration between the two laboratories presents a novel approach in allowing us to simultaneously study the sex-dependent differences at both the individual cellular level and the broader tissue scale. It leverages the specialized expertise of both biomechanics laboratories to offer a multi-dimensional perspective.
2.0 Methods
2.1.1 Tendon Harvest
To begin, dissection kits are autoclaved and 500 mL of both media (Dulbecco’s Modified Eagle Medium, or DMEM, with 10% FBS and 1% PSA) and sterile phosphate-buffered saline (PBS) are made. For one batch of 10 tendons, we prepare the collagenase solution by weighing out 12 mL of collagenase type I, 6 mL of collagenase type IV and place them into a 15 mL conical tube. In the biosafety cabinet (BSC), we add 6 mL of DMEM and pass the solution through a 0.2 μm filter into a new 15 mL tube to sterilize. The tube is parafilmed and placed in a 4℃ refrigerator until ready for digestion. Then, in a 6 well plate, we fill two wells with sterile 1x PBS, one with low glucose sterile DMEM, and one with 70% ethanol. Once complete, the harvest is started.
In order to harvest tendons, we follow the protocol prepared by the Connizzo Lab as follows. A mouse is euthanized and brought into the BSC. The mouse is placed supine on a dissection mat, and then cut halfway down the toes using scissors. Next, we pinch the skin around the knee joint, use a scalpel to create a small cut through the skin at the knee and pull back the skin to the ankle. We then place the foot down on the mat with the anterior side up, score the foot down the middle, and remove the skin around the ankle with coarse forceps or our fingers. We pick the mouse up by its ankle with the bottom of the foot facing towards us, and remove the fat pad with fine forceps until the tendon is visible. We insert our forceps under the tendon and pull away from us to uplift it. Lastly, we pull the tendon towards the medial body of the mouse in order to extract it. When necessary, we cut the fascia connecting the FDL and knee joint first. The tendon is placed in the first PBS well, cleaned with a different set of fine forceps then placed in the second PBS well; the process is repeated for the other leg and 4 other mice.
2.1.2 Digestion
Tenocytes are obtained through digestion. Each clean tendon is quickly dipped in ethanol then DMEM three times each, minced with a blade, and placed in the sterile collagenase solution. This solution is put in a rocker and incubated for 90 to 120 minutes until the tissue is digested. Then, the collagenase solution is quenched with 6 mL of media and filtered through a 70 μm strainer into a 50 mL conical tube. The solution is centrifuged at 750 relative centrifugal field (RCF) for 10 minutes, aspirated, and resuspended in 1 mL media. The cells are seeded into T-25 flasks with an added 5 mL of media and cultured until 80-90% confluency. When they reach this point after about 2 weeks, the cells are passaged. To do this, we aspirate the media, wash the cells twice with sterile 1X PBS (about 3-5 mL), add 3 mL of 0.25% trypsin-EDTA, and let the cells sit in an incubator for up to 5 minutes until all cells are lifted. We quench the trypsin activity by adding 6 mL of media and transfer the cell solution into a new 15-mL tube. Next, we centrifuge at 750 RCF for 10 minutes, aspirate the supernatant, and resuspend the cell pellet in 1 mL of fresh media. After this first passage, we begin experimentation.
2.1.3 Traction Force Microscopy
Materials for the TFM experiment are prepared following the protocol by Bunde et al [20], with few modifications to the precursor To create 13.7 kPa polyacrylamide (PAA) gels as opposed to the published method for a 3.6 kPa gel [20], the PAA hydrogel precursor is prepared using 650 μL of bisacrylamide and 2.44 mL of water. Once the gel is made, it is incubated in 1x PBS at 37 °C for 45 minutes to prepare for cell seeding. Gels are then used immediately in an experiment or stored at 4°C for experiments in the following 24 hours.
The gel is imaged 18 hours later using the imaging software MetaMorph Basic version 7.7.1.0 with a live imaging microscope [22]. The pattern is images using ‘F1 Widefield’ at 40x magnification and cells are imaged using ‘Brightfield Widefield’ illumination at 40x magnification. These respective image series are stacked in a timelapse and then input into the MATLAB code. Next, we enter an elastic modulus of 13600 Pa and set the radius of the dots to 1 μm in the Smith Lab MATLAB code. Finally, we run the code, draw the radius of interest of our cell from the brightfield image in order to tell the code where to look, and analyze the traction force vectors generated by the cells along with the CV. The traction force vectors are calculated according to Equation 1 where E= 13.7 kPa is the Elastic modulus of the gel, a = 1 μm is the patterned dot radius, and v = 0.445 is the gel’s Poisson’s ratio [23].
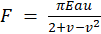
The lab defines T(ti) as the sum of magnitudes of all traction force vectors in the cluster at time t, as shown in Equation 2 where Fk (ti) is the magnitude of the traction force at (xk,ti).
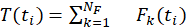
The CV of T(ti) is as described in Equation 3, where σ(T) is the standard deviation [23].
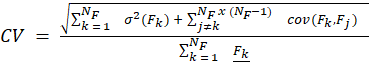
We apply MATLAB’s ttest2 function to compare male and female CVs through a two-sample t-test [24].
2.2.1 Staining Procedures for Bioreactor Experiments
To harvest tendons for bioreactor experiments, we follow the harvest protocol outlined above but keep the flexor tendon explants intact for mechanical testing and imaging.
To prepare imaging, we utilize a staining protocol involving phalloidin and DAPI. Phalloidin staining is performed to visualize the cell membrane, while DAPI staining is used to visualize the cell nuclei. A 400X phalloidin stock solution is prepared by dissolving the vial contents in 150μL of DMSO, yielding a concentration of 2000 assays/mL (~66 μM). 10 μL aliquots of the stock solution are prepared and stored at -20°C until use. The working phalloidin staining solution is created by diluting the stock solution in PBS at a 1:100 ratio. Following this ratio, a working solution of 248 uL PBS and 2.5 uL phalloidin stock is utilized for these experiments. Tendon samples are then incubated in the staining solution for 45 minutes at room temperature in a dark environment, followed by three 5-minute washes with PBS.
After phalloidin staining, we prepare the DAPI stain solution with a dilution ratio of two drops (65 μL) to 1000 μL of PBS is used. The tendon is incubated in this staining solution for a duration of 5 minutes, followed by three 5-minute washes with PBS. The tendon is transferred to a microcentrifuge tube with 1 mL of PBS until imaging is performed.
Once stained, tendon samples are then mounted on glass slides with a drop of PBS to maintain hydration and placed on the microscope stage. Confocal microscopy settings are adjusted accordingly, and the focus is also adjusted in the Z direction to ensure proper visibility of the tendon. The cube turret is set to DAPI and the excitation wavelengths, 405 nm for DAPI and 488 nm for phalloidin, are selected on the laser. Within the FS31V-SV software, the PMT settings are modified as well for best imaging quality– the optimized voltage settings are 750V for DAPI and 585V for phalloidin. Other settings such as laser power can also be adjusted but, once chosen, must remain constant for the duration of the experiment. Continuous use of the High-Low feature on the software while adjusting these settings ensures that the resulting image will not be overexposed. Images are then captured by registering both the low and high ends of the relevant Z range and then beginning the built-in image capture process.
2.2.2 Conducting Bioreactor Experiments
Following staining, tendons are kept in 1 mL of PBS until imaging. The bioreactor is placed into the confocal microscope directly on top of the microscope stage. The attached laptop is turned on, and the MATLAB file Bioreactor App is opened. Ensure the Wi-Fi is connected to allow for seamless experimentation.
To begin loading the tendon for these experiments, the tendon is placed centered on the grip with the foot end of tendon on top of the bottom grip. It is pulled downwards until the edge of the foot is at the edge of the grip. Then, the top grip is secured with two small screws. The other end of the tendon is placed on the second grip and the top grip is loosely secured with two small screws. It is then placed into the grip loader and pulled until the end of the grip loader. Then, all screws of the grip are tightened, and it is placed into the bioreactor within the confocal microscope; 5 mL of PBS are placed on top of the tendon to maintain hydration.
As stated in the staining procedure experiments, confocal microscopy settings are adjusted accordingly. This includes setting the cube turret to DAPI as well as choosing the excitation wavelengths 405 nm for DAPI and 488 nm for phalloidin on the laser instrument. The FS31V-SV software is opened and PMT settings may also be adjusted at this time for best imaging quality. The optimized voltage settings continue to be 750V for DAPI and 585V for phalloidin but other settings under the PMT tab including laser power and the offset may need to be modified to achieve best imaging quality. Use of the High-Low feature throughout the adjustments of the settings will prevent the image from being overexposed. Once the settings are set, a reference image is captured, and the settings are not altered for the duration of the experiment.
To begin experimentation, the MATLAB Bioreactor App is then utilized to move the bioreactor to the home position. The tendon is stretched 5900 um to recreate the prestress that a tendon in a biological system would initially have. Further movements of 100 um increments are executed if the tendon does not appear visibly taut. In the ocular mode of the microscope, the focus is adjusted in the Z direction, and the stage is moved in the X and Y direction to focus on a linear section of the tendon with cells evenly dispersed to allow for tracking. The tendon is then imaged using the built-in method of the microscope by registering both the low and high Z values, creating a Z-stack of images. This image stack will serve as the 0% strain measurement. Using the Bioreactor App again, the tendon is stretched a distance of 300 um with a 100 um/s velocity and 100 um microstep size. The tendon is then relaxed for 10 minutes before the next image is taken. After this period, a new image is captured for the 3% strain measurement. This process of straining the tissue, waiting a ten-minute relaxation period, and imaging the tendon is repeated two more times for the 6% and 9% strain image. The same speed and microstep settings are kept throughout.
2.2.3 Image Processing for Viewing
The images are of the .oib filetype. To create a Z- stack with the images taken with the confocal microscope, the app Fiji ImageJ is required. Using this software, one can open the .oib file and use the Z-stack option to overlay these images. The setting of “maximum intensity” is chosen.
2.2.4 Analyzing Bioreactor Experiment Images
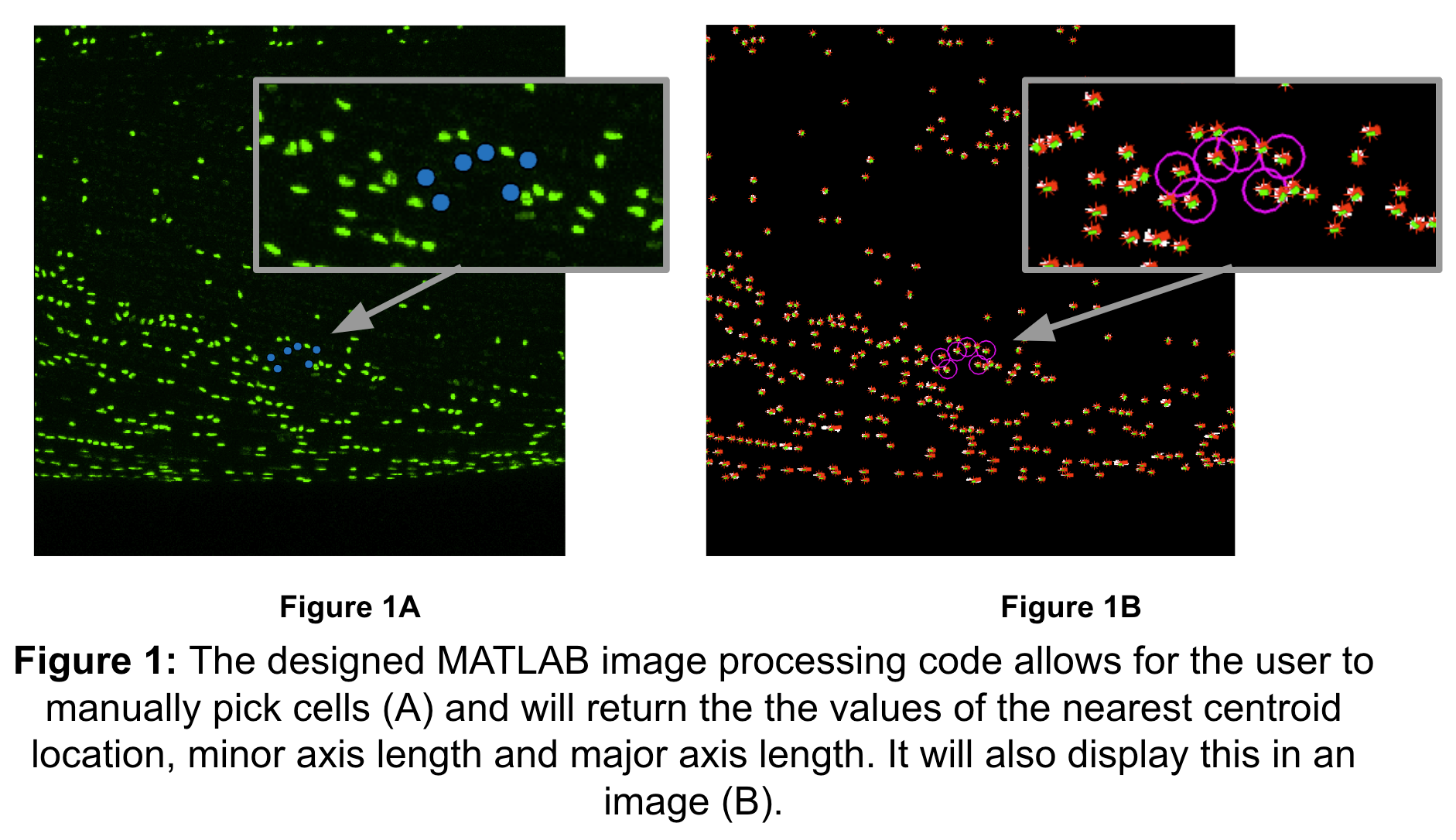
The developed MATLAB data analysis code that is utilized involves the calculation of strain transfer on various length scales to elucidate sex-specific differences in tendon mechanosensing. First, it utilizes the Bio-Formats toolbox to import .oib files and constructs a full Z-stack projection to obtain image projections for DAPI and phalloidin stains. After producing visual representations of these images, it allows for manual selection of individual cell points, binarizes the image, and identifies the nearest cell centroids to the points selected by the user. It also determines the minor and major axes corresponding with each of these centroid values as shown in Figure 1. The next section of the program uses the centroid data to calculate the deformation tensor (F) by dividing the deformed matrix of centroid by the undeformed matrix of centroids, which is used to find the Cauchy-Green deformation tensor (C) through the formula found in Equation 4. Finally, the Lagrangian Strain is calculated following Equation 5. The nuclear strain can be found using Equation 6, where the Ej is the strain in the y-direction and is the second component of the Lagrangian Strain Matrix along the diagonal; Ei is the strain in the x-direction and first component of the Lagrangian Strain Matrix.
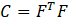
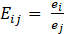
As shown in the mathematical formulas above, the strain tensor derives from the centroids relative to each other over numerous cells; nuclear strain on the other hand is calculated from each individual cell and provides insight into individual cell deformation. After calculating these values, we then perform a two-sample t-test between the male and female nuclear strain values at both 3% and 6% strain using MATLAB’s ttest2 function.
3.0 Results
3.1.1 Tendon Harvest and Cell Culture
Tendon harvests for both TFM and bioreactor experiments were all successful. A total of 96 mouse tendons, 48 female and 48 male, were harvested. Of these tendons, 16 were utilized in bioreactor experiments, and 80 were used in TFM experiments, which were then digested for cell culture. Each digestion contained 10 tendons of the same sex. All 8 digestions were completed successfully as shown through cell culture as each culture flask contained growth of cells within 3-5 days. Of the 8 cell cultures, all cells contained some growth within the first two weeks. One male cell culture became contaminated after 11 days post-digestion. Three other cell cultures, 1 male and 2 female, showed signs of cell death after 3 weeks post-digestion as seen in Figure 2 and eventually died.
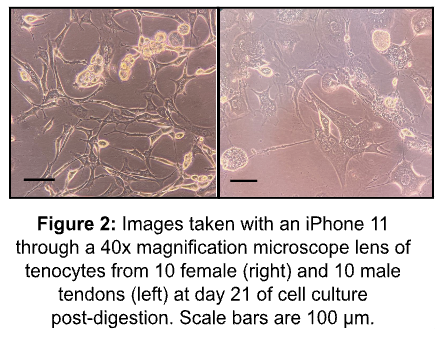
The 4 remaining cell cultures maintained viability with little growth and with no instances of contamination or cell death observed. Two cultures, 1 male and 1 female, grew to around 10% confluency after 3 weeks of cell culture. The other two cultures, 1 male and 1 female, grew to around 10-15% confluency after 2 weeks. Although cells should reach 80-90% confluency 10 days post-digestion, the purposes of this experiment require 65,000 cells per experiment for a viable experiment. Therefore, for this experiment, cell culture was successful in providing an adequate number of cells per TFM experiment.
3.1.2 Traction Force Microscopy Experimentation
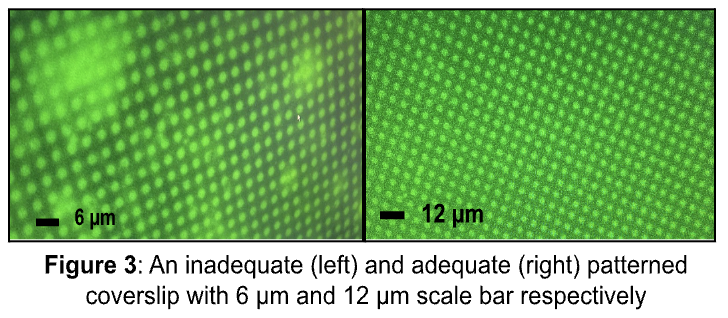
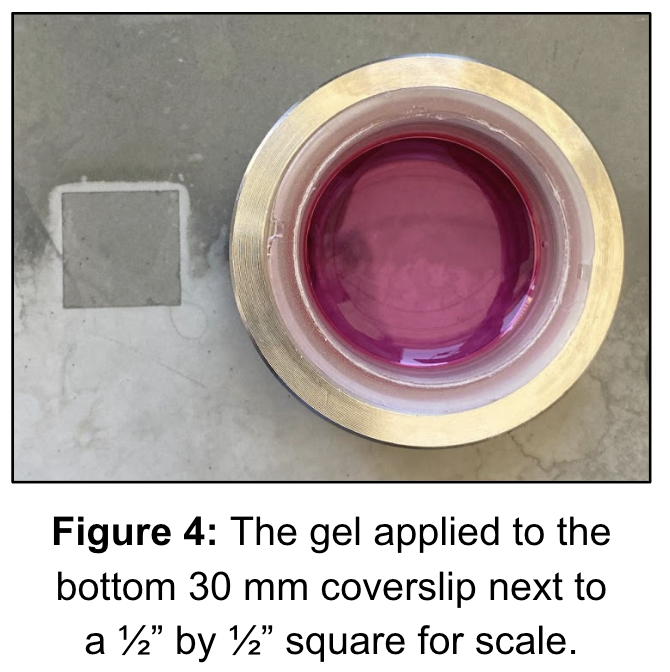
To prepare for final TFM experiments, materials such as top patterned coverslips, bottom coverslips, and PAA gels were created prior to experimentation. The creation of top patterned coverslips were attempted 6 times, each procedure resulting in 7 coverslips. Of the 42 top patterned coverslips created, 6 were found successful to be used in TFM experiments, yielding a 14% success rate. The 35 remaining coverslips were discarded as they were inadequate. Figure 3 illustrates the difference between inadequate coverslips, which show excess protein clumps, and adequate coverslips, which display defined grid micropatterns. 6 bottom coverslips were also cr¬eated, 3 attempts were made and successful. Each attempt yielded 1-3 bottom coverslips to be used in experimentation.
Upon the creation of bottom and top coverslips, PAA gels were created on the 6 bottom coverslips. Figure 4 displays the final assembly of the PAA gel on a treated bottom coverslip, secured by a metal ring set that is screwed together to form a chamber that holds the media. The gel is then seeded with cells. Each time after incubating the cells overnight and imaging them 18 hours later, no live cells or patterns were observed. As shown in Figure 5, the images captured displayed poor results due to unsuccessful pattern transfer onto the gel, which is closely related to cell adherence and survival on the gels.
3.2.1 Staining Procedure
Nuclear staining was successful from the first trial using DAPI, a common live cell stain whose protocols are well-established in standard procedures. To stain the cell membrane, Cell Mask Deep Red (CMDR) was initially utilized. Four trials with varying dilutions and staining durations were tested, as outlined in Table 1, DAPI nuclear stain is displayed in cyan blue, while CMDR membrane stain is displayed as deep blue. Findings from Table 1 indicate that variations in CMDR dilution and incubation time did not yield images with clear cellular staining. Instead, they resulted in excessive background staining and/or non-specific cellular stains.
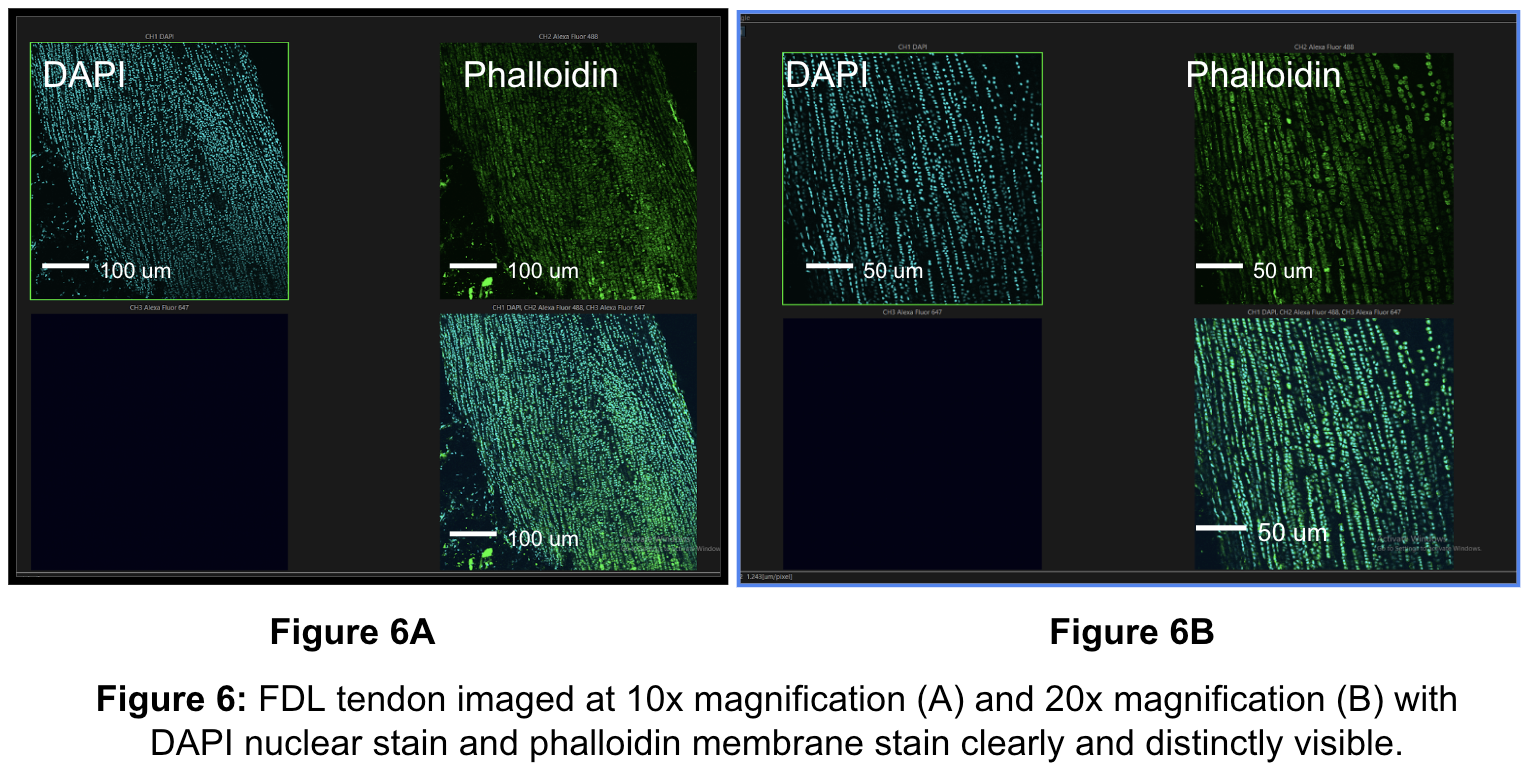
Following the CMDR staining attempts, a new cellular stain was explored. The phalloidin staining protocol described in the methods led to a successful imaging series, as illustrated in Figure 6. Success was constituted by high contrast imaging, distinction of individual cells, and clarity in membrane visualization, all of which are important for cell tracking and strain analysis. The finalized phalloidin and DAPI staining protocols were then used for all of the bioreactor experiments.
3.2.2 Imaging Results
Seven trials were performed using the right and left tendons of four male and female mice. In each experiment, images were acquired for tendons stretched at 0%, 3%, 6%, and 9% strain. Complete and successful data was collected for 11 tendons stained with DAPI and 7 tendons stained with both DAPI and phalloidin. A total of 128 images of data were collected.
Images were acquired using the confocal on the 20x lens with channels for DAPI and phalloidin, as shown in Figure 6. Successful images were characterized by clear tecnocyte morphology, where cells aligned in straight lines indicative of tendon fibers. Additionally, sections were imaged where there was a clear marker that could be tracked across varying strain levels to facilitate accurate cell tracking. Finally, sections where clusters of cells were present did not constitute ideal regions for imaging. Instead, sections where individual cells could be identified constituted ideal imaging regions.
Table 1: CMDR Dilution and Staining Time Trials with Results
|
Trial |
1 |
2 |
3 |
4 |
5 |
|
|
CMDR Dilution |
1:1,000 ul |
1:2,000 ul |
1:5,000 ul |
1:10,000 ul |
1:10,000 ul |
|
|
Staining time |
5 min |
5 min |
5 min |
10 min |
5 min |
|
|
Staining time |
Excess, non-specific background staining. |
Excess, non-specific background staining. |

Section of adipocytes obscuring the target tenocytes. |
|||
3.2.3 Nuclear Strain Calculation and Statistical Analysis
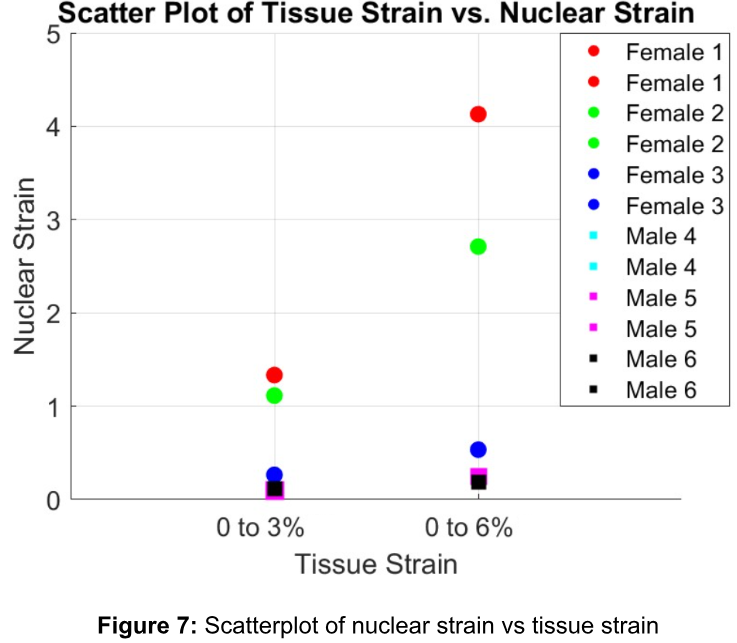
After analyzing the image data acquired from experiments using a MATLAB software described in the methods, the nuclear strain was calculated for each viable trial, resulting in a three-tendon sample size for males and females (a total of six tendons) as shown in Table 2. The analysis focused on the horizontal component (x-direction) of the nuclear strain tensor (ε) due to its alignment with the applied force in the experiment. Consequently, it is anticipated to exhibit the most significant deformation. In Figure 7, the data from Table 2 is plotted on a scatterplot of nuclear strain versus tissue strain. Female trials are represented by circles, while male trials are depicted as squares. Each trial is assigned a unique color for clarity. From this graph, we observe that the horizontal component of the nuclear ε exhibits an increase between the 0-3% and 0-6% tissue strain within the same trial (e.g., female 1), consistent with the anticipated hypothesis of tissue stretching. Additionally, it is important to note that the female groups display greater variation in nuclear strain between trials, whereas the male groups cluster at the bottom of the graph and exhibit very similar nuclear strain across trials. Finally, statistical analysis was performed on the horizontal component of nuclear strain at each tissue strain level (e.g., first within 0-3%, then within 0-6%) to compare the male and female groups. Upon conducting a two-sample t-test, the study found a significant difference between males and females within both the 0 to 3% and 0 to 6% tissue strain groups (p<0.1). Additionally, a two-way ANOVA was conducted and indicated that sex has a significant effect on nuclear strain (p<0.1).
Table 2: Nuclear Strains ε in the X-Direction
|
Strain |
Female |
Male |
||||
|
Nuclear ε 3% (X) |
1.333 |
1.114 |
0.2638 |
0.1201 |
0.09507 |
0.1126 |
|
Nuclear ε 6% (X) |
4.129 |
2.710 |
0.5343 |
0.2401 |
0.2450 |
0.1844 |
4.0 Discussion
4.1 Traction Force Microscopy Experimentation
Results for TFM experiments include harvest, digestion, cell culture, experiment material preparation, imaging of the seeded cells, and data analysis. The harvests and digestion were successful, and cells were cultured. However, cell culture of murine tenocyte exhibited minor success. One culture was contaminated, and three cultures experienced significant cell death after reaching 30-50% confluence, making the cultures unusable for experiments. Contamination most likely occurred due to exposure to unsterile environments, and cell death resulted from inadequate media changes. The other four viable cell cultures showed minimal growth and did not exceed 50% confluency. This lack of growth was likely due to a low seeding density or improper seeding techniques, as cells often grew in small patches. This negatively impacted the potential for successful TFM experiments. Had these issues been resolved, more experiments could have been conducted with better outcomes and data. For future work, we recommend increasing the number of tendons per culture. Using 10 tendons per culture often resulted in a small pellet after digestion. Additionally, ensure sterile environments and frequent media changes to keep cells viable. Future research teams might also consider attempting several dry runs with a non-tenocyte cell line, which may be more robust and easier to work with before attempting experiments with primary cells.
When preparing materials, the top micropatterned coverslips demonstrated a low yet adequate success rate for TFM experiments, requiring multiple trials to obtain a sufficient number of viable coverslips. This is because viable top coverslips require a very high-resolution micropattern. A dark background with well-defined dot separation achieves an optimal signal-to-noise ratio, which is critical for achieving high-resolution imaging and accurately tracking pattern deformation by cells during the experiments. Bottom coverslips were produced with a high success rate, and the protocol was easily repeatable. For future work, we recommend conducting multiple trials to ensure that viable top coverslips are consistently available, thus preventing any delays in the experimental timeline, as these coverslips can be stored indefinitely. Overall, these successful steps enabled us to proceed with making PAA gels. After seeding cells onto the gels and adding media, two sets containing bovine cells leaked, likely due to improper securing or cracking of the bottom coverslip. The other four sets did not leak and were viable for experimentation. During live imaging, no micropatterns or adhered cells were visible, and one gel showed visible debris, likely due to improper pattern transfer onto the gel. To address these issues, we recommend refining the methodologies early with bovine cells, as they are abundant and easier to culture, allowing for extensive experimentation before using murine tenocytes. Additionally, experimenting with higher gel stiffness and different ECM ligands, such as fibronectin or collagen instead of gelatin, may improve cell adherence.
Overall, our results highlight the necessity of prioritizing repeatability. While certain aspects of the protocol were successful, others fell short. Although we successfully prepared all experimental components, live imaging yielded no usable data due to challenges with cell adherence and visualizing patterns. The extended culture time and lack of tenocytes restricted our capacity to conduct multiple experiments with methodological adjustments, emphasizing the importance of beginning with more robust cell types before murine tenocytes.
4.2 Bioreactor Experimentation
Our results substantiate the initial hypothesis that male and female tenocytes exhibit distinct responses under mechanical stress. There was significance in the nuclear strain in the x-direction of 3% and 6% (p < 0.1). Our study's findings are aligned with existing literature that acknowledges sex-dependent variations in cellular and tissue mechanics, such as those documented by Connizzo et al. and Fryhofer et al., which report differential tissue healing capabilities and mechanosensitivity across sexes [18],[19].
Regarding staining procedures, we initially attempted staining FDL tendons with CMDR to visualize cell membranes. The stain was diluted in media to a 1X working solution. We submerged tendon samples in the staining solution for 5 to 10 minutes at room temperature, experimenting with different dilution ratios of CMDR (1:1000, 1:2000, 1:5000, and 1:10000). Despite progressively reducing the CMDR concentration and exploring various washing methods (30-second three-time submersion and 2 minutes of pipetting PBS directly on the tendon), none were successful. Therefore, we explored an alternative method using the phalloidin stain. This new staining protocol facilitated high-contrast imaging and allowed us to distinguish individual cells clearly for subsequent strain analysis and cell tracking. Having now documented and validated the data collection procedure, future efforts can prioritize enhancing data analysis. Our initial trials faced several challenges; in the first trial, PBS was removed to enhance imaging quality, but inadvertently led to excessive tendon shifting and potentially affected nuclear and cellular deformation due to slight tendon dehydration. In the second trial, issues with the Z-range limited data collection to only 0% and 3% strain levels. In the future, precise PBS quantification in the bioreactor well may prevent similar imaging issues. Since the bioreactor was newly machined, we conducted each step of the experiment manually, which was a time-consuming process prone to variability. Future research can work to automate the progressive loading of the tendon and image capturing process. While phalloidin staining was successful in some trials, its effectiveness heavily depended on the quality of the harvested tendon, particularly the thorough removal of fat. Phalloidin preferentially accumulated in fat clumps, causing inconsistent staining and overexposure, rather than an even distribution across the tendon. This highlights the necessity of careful sample preparation to ensure reliable and consistent staining results.
Furthermore, we attempted to manually measure the nuclear deformation using Fiji ImageJ to validate the MATLAB results. However, this method had significant variation due to inaccuracy from manual selection. Thus, we focused on the MATLAB analysis software. Images from successful trials showed cells that could be tracked across images of each strain. Images of the 9% strain were shifted too far beyond the cells captured in the 0% undeformed image, thus there was no reference point to track the cells. We did not calculate cell strain using the phalloidin stain images due to the difficulty of detecting the centroids of the cell membrane images using the MATLAB analysis software. However, the MATLAB analysis software can calculate the cellular strain if the matrices of cell centroids are given as an input.
The data provides quantitative evidence of sex-specific differences in biomechanical response, supporting the hypothesis that male and female tenocytes adapt differently to mechanical cues. Understanding these differences at the cellular and molecular levels can guide the design of more effective treatments for tendon injuries or degenerative diseases. However, potential sources of error may have influenced these results. Variability in tendon handling (including tendon condition, grip pressure, etc.) during the experiments may have led to inconsistent strain application, affecting the measured nuclear strain. Additionally, the limited sample size may not fully capture biological variability, which could skew the interpretation of sex differences. The alignment of the imaging setup and potential artifacts from the imaging process may have also impacted data quality.
Conclusion
The study establishes a protocol for applying mechanical stress on murine tendons for analysis and highlights the need for further experimentation using TFM. The findings indicate a significant difference in nuclear strain between males and females within tissue strain groups and concluded that sex has a significant effect on nuclear strain. While the tissue data showed significance in cell signaling pathways related to ECM homeostasis, further research is recommended to explore sex-based differences, particularly regarding hormonal impact and its role in degenerative diseases. This study provides crucial insights into sex-dependent mechanisms underlying tissue mechanosensing and potential implications for gender-specific therapeutics, encouraging continued research in this area.
Acknowledgements
The authors express their gratitude to Sam Mlawer, Emma Stowe, Allison Sander, Hana Kalco, Matt Lim, Madelyn Keller, Caitlin Colicchio, Anthony Aggouras, and Katie Bunde for their invaluable guidance, support, and training.
References
[1] T. R. Cox and J. T. Erler, "Remodeling and homeostasis of the extracellular matrix: implications for fibrotic diseases and cancer," Disease Models & Mechanisms, vol. 4, no. 2, pp. 165-178, Feb. 2011, doi: https://doi.org/10.1242/dmm.004077. View Article
[2] Z. Peng, H. Sun, V. Bunpetch, Y. Koh, Y. Wen, D. Wu, H. Ouyang, "The regulation of cartilage extracellular matrix homeostasis in joint cartilage degeneration and regeneration," Biomaterials, vol. 268, p. 120555, Jan. 2021, doi: https://doi.org/10.1016/j.biomaterials.2020.120555. View Article
[3] N. Alcorta-Sevillano, I. Macías, A. Infante, and C. I. Rodríguez, "Deciphering the Relevance of Bone ECM Signaling," Cells, vol. 9, no. 12, p. 2630, Dec. 2020, doi: https://doi.org/10.3390/cells9122630. View Article
[4] A. Melouane, M. Yoshioka, and J. St-Amand, "Extracellular matrix/mitochondria pathway: A novel potential target for sarcopenia," Mitochondrion, vol. 50, pp. 63-70, Jan. 2020, doi: https://doi.org/10.1016/j.mito.2019.10.007. View Article
[5] C. Li, N. Wang, A. A. Schaffer, X. Liu, Z. Zhao, G. Elliott, L. Garrett, N. T. Choi, Y. Wang, C. Wang, J. Wang, D. Chan, P. Su, S. Cui, Y. Yang, B. Gao, "Mutations in COMP cause familial carpal tunnel syndrome," Nature Communications, vol. 11, no. 1, Jul. 2020, doi: https://doi.org/10.1038/s41467-020-17378-z. View Article
[6] E. Ortona, M. Pagano, L. Capossela, and W. Malorni, "The Role of Sex Differences in Bone Health and Healing," Biology, vol. 12, no. 7, pp. 993-993, Jul. 2023, doi: https://doi.org/10.3390/biology12070993. View Article
[7] J. Patel, S. Chen, Torey Katzmeyer, Yixuan Amy Pei, and M. Pei, "Sex-dependent variation in cartilage adaptation: from degeneration to regeneration," vol. 14, no. 1, Apr. 2023, doi: https://doi.org/10.1186/s13293-023-00500-3. View Article
[8] Clara Sophie Batzdorf, Anna Sophie Morr, G. Bertalan, I. Sack, Rafaela Vieira Silva, and C. Infante-Duarte, "Sexual Dimorphism in Extracellular Matrix Composition and Viscoelasticity of the Healthy and Inflamed Mouse Brain," Biology, vol. 11, no. 2, pp. 230-230, Jan. 2022, doi: https://doi.org/10.3390/biology11020230. View Article
[9] P. A. Hernandez, M. Moreno, Z. Barati, C. Hutcherson, A. A. Sathe, C. Xing, J. Wright, T. Welch, Y. Dhaher, "Sexual Dimorphism in the Extracellular and Pericellular Matrix of Articular Cartilage," Cartilage, vol. 13, no. 3, p. 194760352211217-194760352211217, Jul. 2022, doi: https://doi.org/10.1177/19476035221121792. View Article
[10] G. Robotti, F. Draghi, C. Bortolotto, and M. G. Canepa, "Ultrasound of sports injuries of the musculoskeletal system: gender differences," Journal of Ultrasound, vol. 23, no. 3, pp. 279-285, Mar. 2020, doi: https://doi.org/10.1007/s40477-020-00438-x. View Article
[11] R. B. Diller and A. J. Tabor, "The Role of the Extracellular Matrix (ECM) in Wound Healing: A Review," Biomimetics, vol. 7, no. 3, p. 87, Jul. 2022, doi: https://doi.org/10.3390/biomimetics7030087. View Article
[12] S. M. Siadat, D. E. Zamboulis, C. T. Thorpe, J. W. Ruberti, and B. K. Connizzo, "Tendon Extracellular Matrix Assembly, Maintenance and Dysregulation Throughout Life," Advances in Experimental Medicine and Biology, pp. 45-103, 2021, doi: https://doi.org/10.1007/978-3-030-80614-9_3. View Article
[13] J. Wertz, M. Galli, and J. R. Borchers, "Achilles Tendon Rupture," Sports Health, vol. 5, no. 5, pp. 407-409, Sep. 2013, doi: https://doi.org/10.1177/1941738112472165. View Article
[14] L. I. Pease, P. D. Clegg, C. J. Proctor, D. J. Shanley, S. J. Cockell, and M. J. Peffers, "Cross platform analysis of transcriptomic data identifies ageing has distinct and opposite effects on tendon in males and females," Scientific Reports, vol. 7, no. 1, Oct. 2017, doi: https://doi.org/10.1038/s41598-017-14650-z. View Article
[15] J. Buschmann and G. Meier Bürgisser, "Structure and function of tendon and ligament tissues," Biomechanics of Tendons and Ligaments, pp. 3-29, 2017, doi: https://doi.org/10.1016/b978-0-08-100489-0.00001-6. View Article
[16] E. Maeda, M. Sugimoto, and T. Ohashi, "Cytoskeletal tension modulates MMP-1 gene expression from tenocytes on micropillar substrates," Journal of Biomechanics, vol. 46, no. 5, pp. 991-997, Mar. 2013, doi: https://doi.org/10.1016/j.jbiomech.2012.11.056. View Article
[17] M. E. Wall, P. S. Weinhold, T. Siu, T. Brown, and A. J. Banes, "Comparison of cellular strain with applied substrate strain in vitro," Journal of Biomechanics, vol. 40, no. 1, pp. 173-181, Jan. 2007, doi: https://doi.org/10.1016/j.jbiomech.2005.10.032. View Article
[18] B. K. Connizzo, J. M. Piet, S. J. Shefelbine, and A. J. Grodzinsky, "Age-associated changes in the response of tendon explants to stress deprivation is sex-dependent," Connective Tissue Research, vol. 61, no. 1, pp. 48-62, Aug. 2019, doi: https://doi.org/10.1080/03008207.2019.1648444. View Article
[19] G. W. Fryhofer, B. R. Freedman, C. D. Hillin, N. S. Salka, A. M. Pardes, S. N. Weiss, D. C. Farber, and L. J. Soslowsky, "Postinjury biomechanics of Achilles tendon vary by sex and hormone status," Journal of Applied Physiology, vol. 121, no. 5, pp. 1106-1114, Nov. 2016, doi: https://doi.org/10.1152/japplphysiol.00620.2016. View Article
[20] K. A. Bunde, D. Stamenović, and M. L. Smith, "Pattern Generation for Micropattern Traction Microscopy," Journal of visualized experiments, no. 180, Feb. 2022, doi: https://doi.org/10.3791/63628. View Article
[21] J. Li, P. E. Barbone, M. L. Smith, and Dimitrije Stamenović, "Effect of correlation between traction forces on tensional homeostasis in clusters of endothelial cells and fibroblasts," Journal of biomechanics, vol. 100, pp. 109588-109588, Feb. 2020, doi: https://doi.org/10.1016/j.jbiomech.2019.109588. View Article
[22] "MetaMorph Microscopy Automation and Image Analysis Software," www.moleculardevices.com. (accessed May 29, 2024). View Article
[23] H. Xu, S. Donegan, J. M. Dreher, A. J. Stark, E. P. Canović, D. Stamenović, M. L. Smith, "Focal adhesion displacement magnitude is a unifying feature of tensional homeostasis," Acta biomaterialia, vol. 113, pp. 372-379, Sep. 2020, doi: https://doi.org/10.1016/j.actbio.2020.06.043. View Article
[24] "Two-sample t-test - MATLAB ttest2," www.mathworks.com. View Article

